Ecological Trajectory Analysis (ETA) is a framework to analyze dynamics of ecosystems described as trajectories in a chosen space of multivariate resemblance (De Cáceres et al. 2019). ETA takes trajectories as objects to be analyzed and compared geometrically.
Usage
segmentDistances(
d,
sites,
surveys = NULL,
distance.type = "directed-segment",
add = TRUE,
verbose = FALSE
)
trajectoryDistances(
d,
sites,
surveys = NULL,
distance.type = "DSPD",
symmetrization = "mean",
add = TRUE,
verbose = FALSE
)
trajectoryLengths(
d,
sites,
surveys = NULL,
relativeToInitial = FALSE,
all = FALSE,
verbose = FALSE
)
trajectoryLengths2D(
xy,
sites,
surveys,
relativeToInitial = FALSE,
all = FALSE,
verbose = FALSE
)
trajectoryAngles(
d,
sites,
surveys = NULL,
all = FALSE,
relativeToInitial = FALSE,
stats = TRUE,
add = TRUE,
verbose = FALSE
)
trajectoryAngles2D(
xy,
sites,
surveys,
relativeToInitial = FALSE,
betweenSegments = TRUE
)
trajectoryProjection(d, target, trajectory, tol = 1e-06, add = TRUE)
trajectoryConvergence(
d,
sites,
surveys = NULL,
symmetric = FALSE,
add = TRUE,
verbose = FALSE
)
trajectoryDirectionality(d, sites, surveys = NULL, add = TRUE, verbose = FALSE)Arguments
- d
A symmetric
matrixor an object of classdistcontaining the distance values between pairs of ecosystem states (see details).- sites
A vector indicating the site corresponding to each ecosystem state.
- surveys
A vector indicating the survey corresponding to each ecosystem state (only necessary when surveys are not in order).
- distance.type
The type of distance index to be calculated (Besse et al. 2016; De Cáceres et al. submitted). For
segmentDistancesthe available indices are:Hausdorff: Hausdorff distance between two segments.directed-segment: Directed segment distance (default).PPA: Perpendicular-parallel-angle distance.
whereas for
trajectoryDistancesthe available indices are:Hausdorff: Hausdorff distance between two trajectories.SPD: Segment path distance.DSPD: Directed segment path distance (default).
- add
Flag to indicate that constant values should be added (local transformation) to correct triplets of distance values that do not fulfill the triangle inequality.
- verbose
Provides console output informing about process (useful for large dataset).
- symmetrization
Function used to obtain a symmetric distance, so that DSPD(T1,T2) = DSPD(T2,T1) (e.g.,
meanormin). Ifsymmetrization = NULLthen the symmetrization is not conducted and the output dissimilarity matrix is not symmetric.- relativeToInitial
Flag to indicate that lengths or angles should be calculated with respect to initial survey.
- all
A flag to indicate that angles are desired for all triangles (i.e. all pairs of segments) in the trajectory. If FALSE, angles are calculated for consecutive segments only.
- xy
Matrix with 2D coordinates in a Cartesian space (typically an ordination of ecosystem states).
- stats
A flag to indicate that circular statistics are desired (mean, standard deviation and mean resultant length, i.e. rho)
- betweenSegments
Flag to indicate that angles should be calculated between trajectory segments or with respect to X axis.
- target
An integer vector of the ecosystem states to be projected.
- trajectory
An integer vector of the trajectory onto which target states are to be projected.
- tol
Numerical tolerance value to determine that projection of a point lies within the trajectory.
- symmetric
A logical flag to indicate a symmetric convergence comparison of trajectories.
Value
Function trajectoryDistances returns an object of class dist containing the distances between trajectories (if symmetrization = NULL then the object returned is of class matrix).
Function trajectorySegments returns a list with the following elements:
Dseg: Distance matrix between segments.Dini: Distance matrix between initial points of segments.Dfin: Distance matrix between final points of segments.Dinifin: Distance matrix between initial points of one segment and the final point of the other.Dfinini: Distance matrix between final points of one segment and the initial point of the other.
Function trajectoryLengths returns a data frame with the length of each segment on each trajectory and the total length of all trajectories.
If relativeToInitial = TRUE lengths are calculated between the initial survey and all the other surveys.
If all = TRUE lengths are calculated for all segments.
Function trajectoryLengths2D returns a data frame with the length of each segment on each trajectory and the total length of all trajectories.
If relativeToInitial = TRUE lengths are calculated between the initial survey and all the other surveys.
If all = TRUE lengths are calculated for all segments.
Function trajectoryAngles returns a data frame with angle values on each trajectory. If stats=TRUE, then the mean, standard deviation and mean resultant length of those angles are also returned.
Function trajectoryAngles2D returns a data frame with angle values on each trajectory. If betweenSegments=TRUE, then angles are calculated between trajectory segments, alternatively, If betweenSegments=FALSE, angles are calculated considering Y axis as the North (0°).
Function trajectoryProjection returns a data frame with the following columns:
distanceToTrajectory: Distances to the trajectory, i.e. rejection (NAfor target points whose projection is outside the trajectory).segment: Segment that includes the projected point (NAfor target points whose projection is outside the trajectory).relativePosition: Relative position of the projected point within the trajectory, i.e. values from 0 to 1 with 0 representing the start of the trajectory and 1 representing the end (NAfor target points whose projection is outside the trajectory).
Function trajectoryConvergence returns a list with two elements:
tau: A matrix with the statistic (Mann-Kendall's tau) of the convergence/divergence test between trajectories. Ifsymmetric=TRUEthen the matrix is square. Otherwise the statistic of the test of the row trajectory approaching the column trajectory.p.value: A matrix with the p-value of the convergence/divergence test between trajectories. Ifsymmetric=TRUEthen the matrix is square. Otherwise the p-value indicates the test of the row trajectory approaching the column trajectory.
Function trajectoryDirectionality returns a vector with directionality values (one per trajectory).
Details
Given a distance matrix between ecosystem states, the set of functions that provide ETA metrics are:
Functions
segmentDistancesandtrajectoryDistancescalculate the distance between pairs of directed segments and ecosystem trajectories, respectively.Function
trajectoryLengthscalculates lengths of directed segments and total path lengths of trajectories.Function
trajectoryLengths2Dcalculates lengths of directed segments and total path lengths of trajectories from 2D coordinates given as input.Function
trajectoryAnglescalculates the angle between consecutive pairs of directed segments or between segments of ordered triplets of points.Function
trajectoryAngles2Dcalculates the angle between consecutive pairs of directed segments or between segments of ordered triplets of points.Function
trajectoryProjectionprojects a set of target points onto a specified trajectory and returns the distance to the trajectory (i.e. rejection) and the relative position of the projection point within the trajectory.Function
trajectoryConvergenceperforms the Mann-Kendall trend test on the distances between trajectories (symmetric test) or the distance between points of one trajectory to the other.Function
trajectoryDirectionalityreturns (for each trajectory) a statistic that measures directionality of the whole trajectory.
Details of calculations are given in De Cáceres et al (2019).
The input distance matrix d should ideally be metric. That is, all subsets of distance triplets should fulfill the triangle inequality (see utility function is.metric).
All ETA functions that require metricity include a parameter 'add', which by default is TRUE, meaning that whenever the triangle inequality is broken the minimum constant required to fulfill it is added to the three distances.
If such local (an hence, inconsistent across triplets) corrections are not desired, users should find another way modify d to achieve metricity, such as PCoA, metric MDS or non-metric MDS (see vignette 'Introduction to Ecological Trajectory Analysis').
If parameter 'add' is set to FALSE and problems of triangle inequality exist, ETA functions may provide missing values in some cases where they should not.
The resemblance between trajectories is done by adapting concepts and procedures used for the analysis of trajectories in space (i.e. movement data) (Besse et al. 2016).
Function trajectoryAngles calculates angles between consecutive segments in degrees. For each pair of segments, the angle between the two is defined on the plane that contains the two segments, and measures the change in direction (in degrees) from one segment to the other.
Angles are always positive, with zero values indicating segments that are in a straight line, and values equal to 180 degrees for segments that are in opposite directions. If all = TRUE
angles are calculated between the segments corresponding to all ordered triplets. Alternatively, if relativeToInitial = TRUE angles are calculated for each segment with respect to the initial survey.
Function trajectoryAngles2D calculates angles between consecutive segments in degrees from 2D coordinates given as input. For each pair of segments, the angle between the two is defined on the plane that contains the two segments, and measures the change in direction (in degrees) from one segment to the other.
Angles are always positive (O to 360), with zero values indicating segments that are in a straight line, and values equal to 180 degrees for segments that are in opposite directions.
If all = TRUE angles are calculated between the segments corresponding to all ordered triplets. Alternatively, if relativeToInitial = TRUE angles are calculated for each segment with respect to the initial survey.
If betweenSegments = TRUE angles are calculated between segments of trajectory, otherwise, If betweenSegments = FALSE, angles are calculated considering Y axis as the North (0°).
References
Besse, P., Guillouet, B., Loubes, J.-M. & François, R. (2016). Review and perspective for distance based trajectory clustering. IEEE Trans. Intell. Transp. Syst., 17, 3306–3317.
De Cáceres M, Coll L, Legendre P, Allen RB, Wiser SK, Fortin MJ, Condit R & Hubbell S. (2019). Trajectory analysis in community ecology. Ecological Monographs 89, e01350.
Author
Miquel De Cáceres, CREAF
Anthony Sturbois, Vivarmor nature, Réserve Naturelle nationale de la Baie de Saint-Brieuc
Examples
#Description of sites and surveys
sites = c(1,1,1,2,2,2)
surveys=c(1,2,3,1,2,3)
#Raw data table
xy<-matrix(0, nrow=6, ncol=2)
xy[2,2]<-1
xy[3,2]<-2
xy[4:6,1] <- 0.5
xy[4:6,2] <- xy[1:3,2]
xy[6,1]<-1
#Draw trajectories
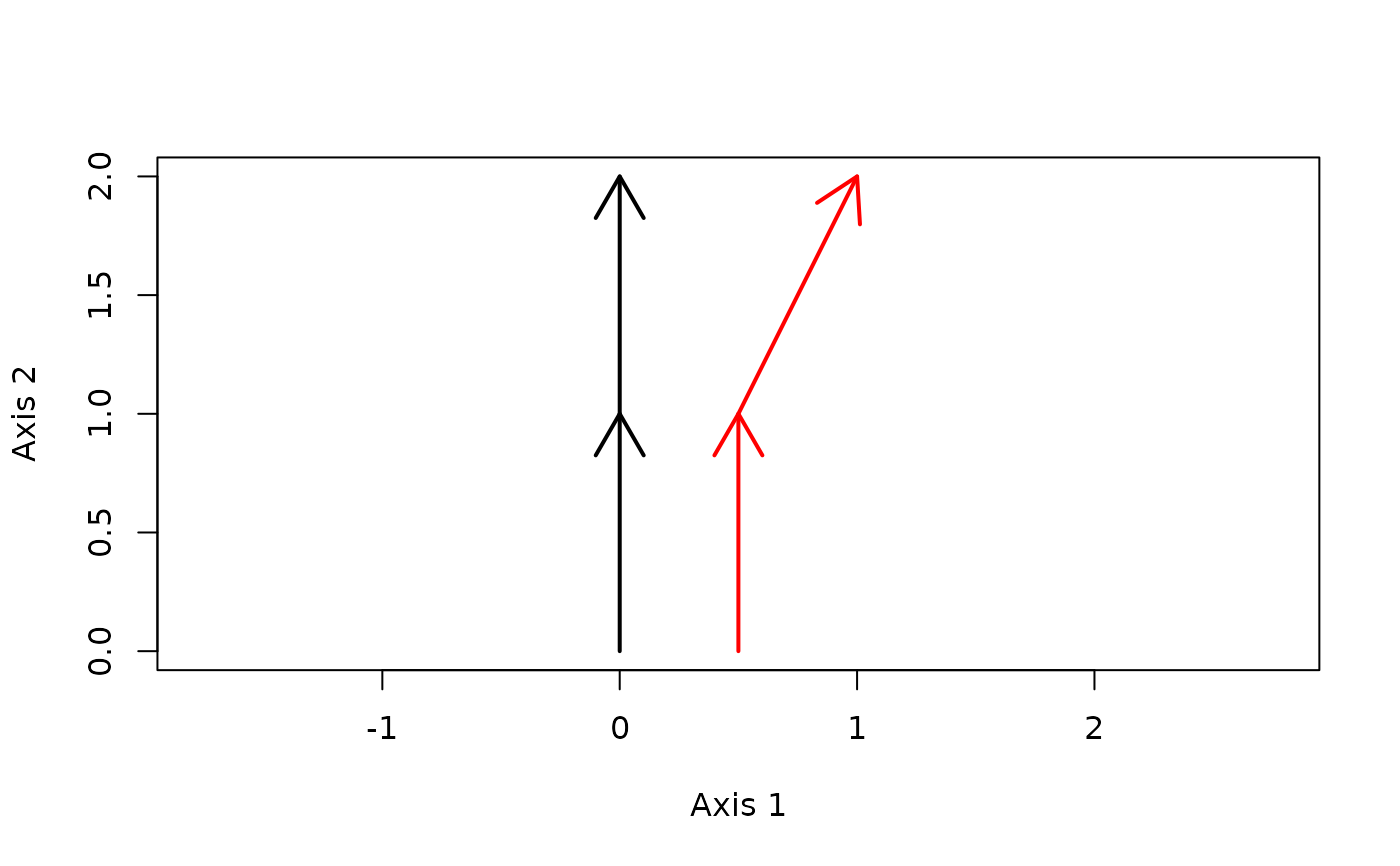
trajectoryPlot(xy, sites, surveys,
traj.colors = c("black","red"), lwd = 2)
 #Distance matrix
d = dist(xy)
d
#> 1 2 3 4 5
#> 2 1.000000
#> 3 2.000000 1.000000
#> 4 0.500000 1.118034 2.061553
#> 5 1.118034 0.500000 1.118034 1.000000
#> 6 2.236068 1.414214 1.000000 2.061553 1.118034
trajectoryLengths(d, sites, surveys)
#> S1 S2 Trajectory
#> 1 1 1.000000 2.000000
#> 2 1 1.118034 2.118034
trajectoryLengths2D(xy, sites, surveys)
#> S1 S2 Trajectory
#> 1 1 1.000000 2.000000
#> 2 1 1.118034 2.118034
trajectoryAngles(d, sites, surveys)
#> S1-S2 mean sd rho
#> 1 0.00000 0.00000 0.000000e+00 1
#> 2 26.56505 26.56505 1.490116e-08 1
trajectoryAngles2D(xy, sites, surveys, betweenSegments = TRUE)
#> t1-t2
#> 1 0
#> 2 26.565051177078
trajectoryAngles2D(xy, sites, surveys, betweenSegments = FALSE)
#> Axis2-t1 Axis2-t2
#> 1 0 0.00000
#> 2 0 26.56505
segmentDistances(d, sites, surveys)$Dseg
#> 1[1-2] 1[2-3] 2[1-2]
#> 1[2-3] 1.000000
#> 2[1-2] 0.500000 1.118034
#> 2[2-3] 1.414214 1.000000 1.118034
trajectoryDistances(d, sites, surveys, distance.type = "Hausdorff")
#> 1
#> 2 1.414214
trajectoryDistances(d, sites, surveys, distance.type = "DSPD")
#> 1
#> 2 0.75
#Should give the same results if surveys are not in order
#(here we switch surveys for site 2)
temp = xy[5,]
xy[5,] = xy[6,]
xy[6,] = temp
surveys[5] = 3
surveys[6] = 2
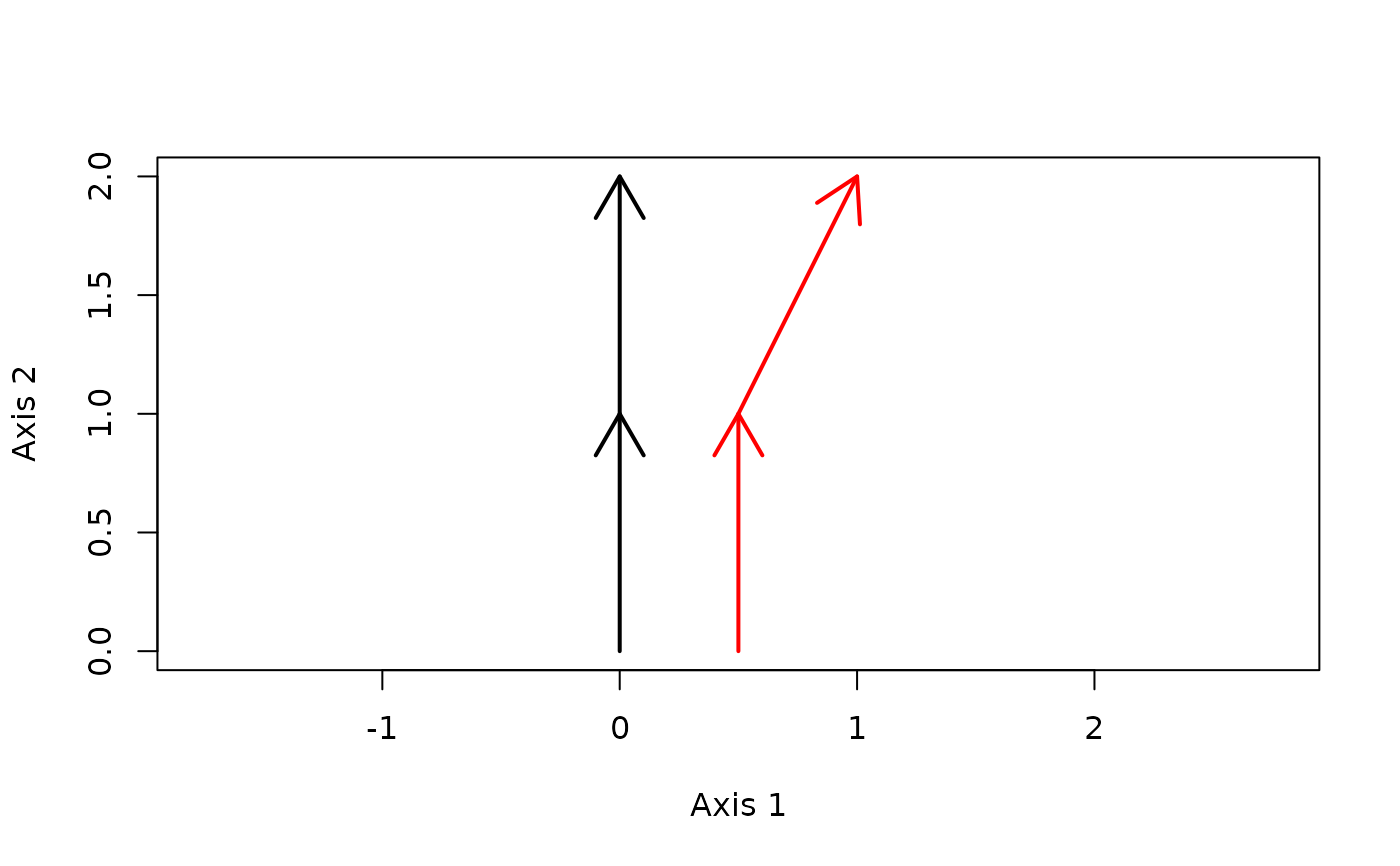
trajectoryPlot(xy, sites, surveys,
traj.colors = c("black","red"), lwd = 2)
#Distance matrix
d = dist(xy)
d
#> 1 2 3 4 5
#> 2 1.000000
#> 3 2.000000 1.000000
#> 4 0.500000 1.118034 2.061553
#> 5 1.118034 0.500000 1.118034 1.000000
#> 6 2.236068 1.414214 1.000000 2.061553 1.118034
trajectoryLengths(d, sites, surveys)
#> S1 S2 Trajectory
#> 1 1 1.000000 2.000000
#> 2 1 1.118034 2.118034
trajectoryLengths2D(xy, sites, surveys)
#> S1 S2 Trajectory
#> 1 1 1.000000 2.000000
#> 2 1 1.118034 2.118034
trajectoryAngles(d, sites, surveys)
#> S1-S2 mean sd rho
#> 1 0.00000 0.00000 0.000000e+00 1
#> 2 26.56505 26.56505 1.490116e-08 1
trajectoryAngles2D(xy, sites, surveys, betweenSegments = TRUE)
#> t1-t2
#> 1 0
#> 2 26.565051177078
trajectoryAngles2D(xy, sites, surveys, betweenSegments = FALSE)
#> Axis2-t1 Axis2-t2
#> 1 0 0.00000
#> 2 0 26.56505
segmentDistances(d, sites, surveys)$Dseg
#> 1[1-2] 1[2-3] 2[1-2]
#> 1[2-3] 1.000000
#> 2[1-2] 0.500000 1.118034
#> 2[2-3] 1.414214 1.000000 1.118034
trajectoryDistances(d, sites, surveys, distance.type = "Hausdorff")
#> 1
#> 2 1.414214
trajectoryDistances(d, sites, surveys, distance.type = "DSPD")
#> 1
#> 2 0.75
#Should give the same results if surveys are not in order
#(here we switch surveys for site 2)
temp = xy[5,]
xy[5,] = xy[6,]
xy[6,] = temp
surveys[5] = 3
surveys[6] = 2
trajectoryPlot(xy, sites, surveys,
traj.colors = c("black","red"), lwd = 2)
 trajectoryLengths(dist(xy), sites, surveys)
#> S1 S2 Trajectory
#> 1 1 1.000000 2.000000
#> 2 1 1.118034 2.118034
trajectoryLengths2D(xy, sites, surveys)
#> S1 S2 Trajectory
#> 1 1 1.000000 2.000000
#> 2 1 1.118034 2.118034
segmentDistances(dist(xy), sites, surveys)$Dseg
#> 1[1-2] 1[2-3] 2[1-2]
#> 1[2-3] 1.000000
#> 2[1-2] 0.500000 1.118034
#> 2[2-3] 1.414214 1.000000 1.118034
trajectoryAngles(dist(xy), sites, surveys)
#> S1-S2 mean sd rho
#> 1 0.00000 0.00000 0.000000e+00 1
#> 2 26.56505 26.56505 1.490116e-08 1
trajectoryAngles2D(xy, sites, surveys, betweenSegments = TRUE)
#> t1-t2
#> 1 0
#> 2 26.565051177078
trajectoryAngles2D(xy, sites, surveys, betweenSegments = FALSE)
#> Axis2-t1 Axis2-t2
#> 1 0 0.00000
#> 2 0 26.56505
trajectoryDistances(dist(xy), sites, surveys, distance.type = "Hausdorff")
#> 1
#> 2 1.414214
trajectoryDistances(dist(xy), sites, surveys, distance.type = "DSPD")
#> 1
#> 2 0.75
trajectoryLengths(dist(xy), sites, surveys)
#> S1 S2 Trajectory
#> 1 1 1.000000 2.000000
#> 2 1 1.118034 2.118034
trajectoryLengths2D(xy, sites, surveys)
#> S1 S2 Trajectory
#> 1 1 1.000000 2.000000
#> 2 1 1.118034 2.118034
segmentDistances(dist(xy), sites, surveys)$Dseg
#> 1[1-2] 1[2-3] 2[1-2]
#> 1[2-3] 1.000000
#> 2[1-2] 0.500000 1.118034
#> 2[2-3] 1.414214 1.000000 1.118034
trajectoryAngles(dist(xy), sites, surveys)
#> S1-S2 mean sd rho
#> 1 0.00000 0.00000 0.000000e+00 1
#> 2 26.56505 26.56505 1.490116e-08 1
trajectoryAngles2D(xy, sites, surveys, betweenSegments = TRUE)
#> t1-t2
#> 1 0
#> 2 26.565051177078
trajectoryAngles2D(xy, sites, surveys, betweenSegments = FALSE)
#> Axis2-t1 Axis2-t2
#> 1 0 0.00000
#> 2 0 26.56505
trajectoryDistances(dist(xy), sites, surveys, distance.type = "Hausdorff")
#> 1
#> 2 1.414214
trajectoryDistances(dist(xy), sites, surveys, distance.type = "DSPD")
#> 1
#> 2 0.75