Function coverage calculates the proportion of sites of the target site group where one or another indicator (a species or a species combination) is found. Parameters are used to select valid indicators before calculating coverage. Function plotcoverage plots the coverage against the positive predictive value threshold used to select indicators.
Usage
coverage(
x,
y = NULL,
selection = NULL,
minstat = NULL,
At = NULL,
Bt = NULL,
type = "stat",
alpha = NULL
)
plotcoverage(
x,
y = NULL,
by = 0.05,
type = "stat",
max.order = NULL,
group = NULL,
alpha = NULL,
add = FALSE,
xlab = expression(A[t]),
...
)Arguments
- x
An object of class '
indicators' or a community data table with sites in rows and species in columns (in this second case, an object of class 'multipatt' must be supplied fory).- y
An object of class '
multipatt'.- selection
A logical vector restricting the set of indicators used to calculate the coverage.
- minstat
Minimum value of the statistic for selecting indicators.
- At
Minimum value of positive predictive value (A) for selecting indicators.
- Bt
Minimum value for sensitivity (B) for selecting indicators.
- type
Specifies how to select indicators: (1) using the value of the statistic (
type = "stat"); (2) the lower bound of its confidence interval (type = "lowerCI"); or (3) the upper bound of its confidence interval (type = "upperCI"). This parameter makes sense when the function is called using objects of class 'indicators' and bootstrap confidence intervals are available for this object. Otherwisetypehas no effect and the value of the statistic is used for selection. In functioncoverage, the value oftypeapplies to selection usingminstat,AtandBt. In functionplotcoverage, the value oftypeapplies to selection usingAt.- alpha
Significance level for selecting indicators.
- by
Rate of increase in the predictive value threshold (At).
- max.order
The maximum number of species conforming species combinations (allows examining the effects of increasing the order of combinations).This parameter is only used when the function is called using objects of class '
indicators'.- group
Either an integer or a character string indicating the site group or site group combination for which plot is desired. This parameter is only used when the function is called using objects of class '
multipatt'.- add
Flag indicating whether the function should draw on the existing plot.
- xlab
Label for the x-axis.
- ...
Additional plotting parameters that are passed to the
plotfunction.
Value
When used with an object of class 'indicators', function coverage returns the proportion of sites of the target site group where one or another indicator (species combination) is found. When used with an object of class 'indicators', function coverage returns a vector containing the coverage value for each site group or site group combination.
Details
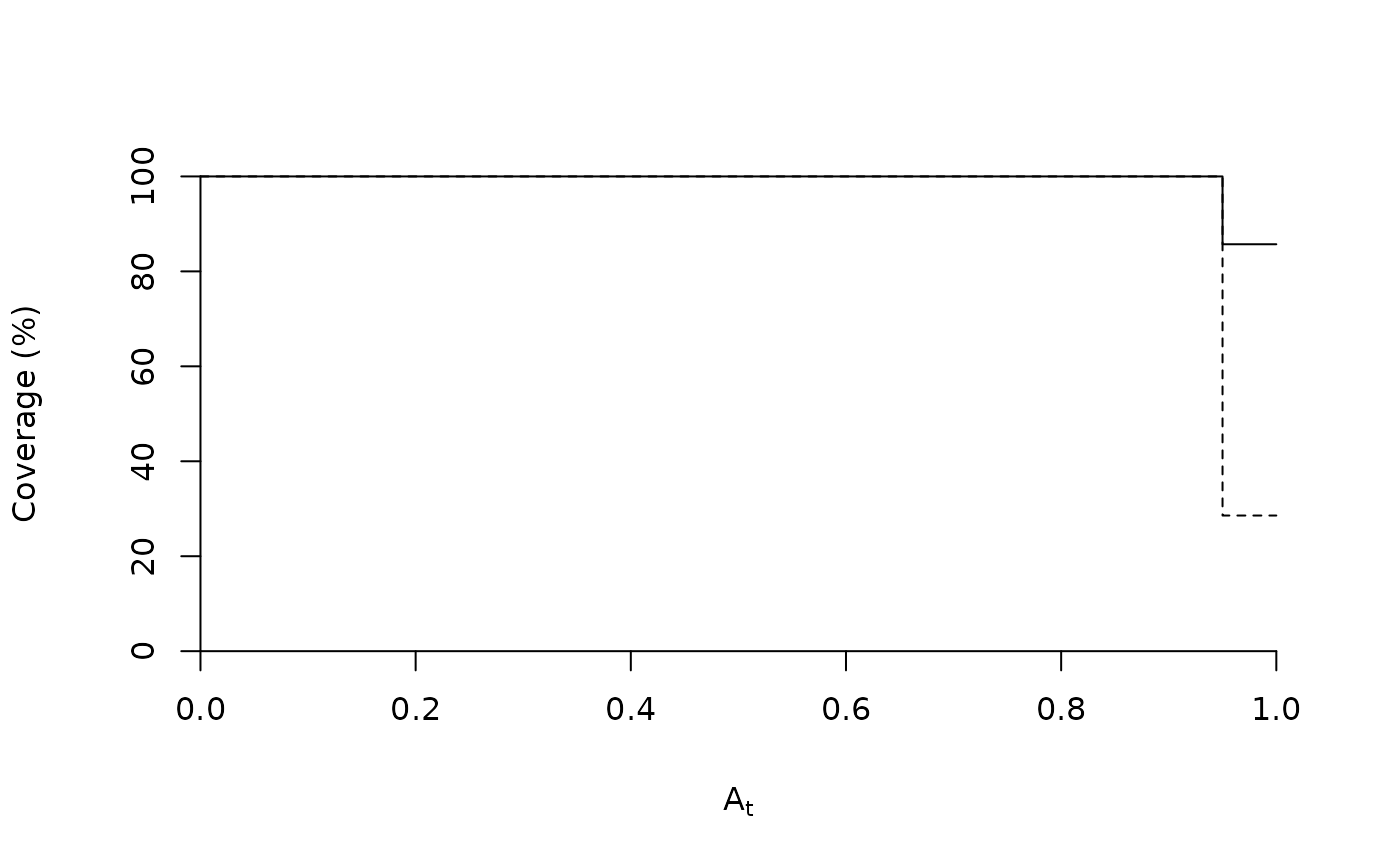
The coverage of a set of indicators was defined in De Cáceres et al. (2012) as the proportion of sites in a given site group where one or several indicators are found. This value allows assessing how often the site group will be able to be determined. If all indicators of a site group are rare, then the indication system will not be useful, regardless of how much restricted to the site group the indicators are. The coverage value is a generalization of quantity B of IndVal, that applies to a group of indicators instead of a single one. Function plotcoverage plots the coverage against the positive predictive value threshold (At) used to select indicators, as in De Cáceres et al. (2012). Functions coverage and plotcoverage can be executed using either an object of class 'indicators', or an object of class 'multipatt'. However, the parameters that apply to each case are slightly different. When using coverage and plotcoverage on objects of class 'multipatt' one is expected to calculate the coverage for those indicators that are significant (see alpha parameter), although other constraints to select valid indicators can be used. When using coverage and plotcoverage on objects of class 'indicators' one is expected to calculate the coverage for indicators that have values of A larger than a specified threshold (see At parameter). In this latter case, it may be advisable to use stat="lowerCI", so that indicators with broad confidence intervals are not included in the selection.
References
De Cáceres, M., Legendre, P., Wiser, S.K. and Brotons, L. 2012. Using species combinations in indicator analyses. Methods in Ecology and Evolution 3(6): 973-982.
Examples
library(stats)
data(wetland) ## Loads species data
## Creates three clusters using kmeans
wetkm <- kmeans(wetland, centers=3)
## Run indicator analysis with species combinations for the first group
sc <- indicators(X=wetland, cluster=wetkm$cluster, group=1, verbose=TRUE, At=0.5, Bt=0.2)
#> Target site group: 1
#> Number of candidate species: 33
#> Number of sites: 41
#> Size of the site group: 10
#> Starting species 1 ... accepted combinations: 0
#> Starting species 2 ... accepted combinations: 16
#> Starting species 3 ... accepted combinations: 16
#> Starting species 4 ... accepted combinations: 87
#> Starting species 5 ... accepted combinations: 87
#> Starting species 6 ... accepted combinations: 133
#> Starting species 7 ... accepted combinations: 133
#> Starting species 8 ... accepted combinations: 153
#> Starting species 9 ... accepted combinations: 155
#> Starting species 10 ... accepted combinations: 155
#> Starting species 11 ... accepted combinations: 155
#> Starting species 12 ... accepted combinations: 155
#> Starting species 13 ... accepted combinations: 155
#> Starting species 14 ... accepted combinations: 155
#> Starting species 15 ... accepted combinations: 155
#> Starting species 16 ... accepted combinations: 160
#> Starting species 17 ... accepted combinations: 161
#> Starting species 18 ... accepted combinations: 170
#> Starting species 19 ... accepted combinations: 170
#> Starting species 20 ... accepted combinations: 172
#> Starting species 21 ... accepted combinations: 172
#> Starting species 22 ... accepted combinations: 173
#> Starting species 23 ... accepted combinations: 173
#> Starting species 24 ... accepted combinations: 173
#> Starting species 25 ... accepted combinations: 173
#> Starting species 26 ... accepted combinations: 173
#> Starting species 27 ... accepted combinations: 173
#> Starting species 28 ... accepted combinations: 173
#> Starting species 29 ... accepted combinations: 173
#> Starting species 30 ... accepted combinations: 173
#> Starting species 31 ... accepted combinations: 173
#> Starting species 32 ... accepted combinations: 173
#> Starting species 33 ... accepted combinations: 173
#> Number of valid combinations: 173
#> Number of remaining species: 13
#> Calculating statistical significance (permutational test)...
## Determine the coverage of the selected set of indicators
coverage(sc)
#> [1] 1
## Plot the coverage against the threshold At
plotcoverage(sc)
plotcoverage(sc, max.order=2, add=TRUE, lty=2)
 ## Runs the combination analysis using IndVal.g as statistic
wetpt <- multipatt(wetland, wetkm$cluster, control = how(nperm=999))
## Determines the coverage for each site group combination
coverage(wetland, wetpt, alpha = 0.05)
#> 1 2 3 1+2 1+3 2+3 1+2+3
#> 1.0000000 1.0000000 1.0000000 0.8750000 0.0000000 0.7741935 0.0000000
## Runs the combination analysis using IndVal.g as statistic
wetpt <- multipatt(wetland, wetkm$cluster, control = how(nperm=999))
## Determines the coverage for each site group combination
coverage(wetland, wetpt, alpha = 0.05)
#> 1 2 3 1+2 1+3 2+3 1+2+3
#> 1.0000000 1.0000000 1.0000000 0.8750000 0.0000000 0.7741935 0.0000000