
Forest dynamics
Miquel De Caceres
2026-02-27
Source:vignettes/runmodels/ForestDynamics.Rmd
ForestDynamics.RmdAbout this vignette
This document describes how to run the forest dynamics model of
medfate, described in De Cáceres et al. (2023) and
implemented in function fordyn(). This document is meant to
teach users to run the simulation model with function
fordyn(). Details of the model design and formulation can
be found at the corresponding chapters of the medfate
book.
Because the model builds on the growth and water balance models, the
reader is assumed here to be familiarized with spwb() and
growth() (otherwise read vignettes Basic
water balance and Forest
growth).
Preparing model inputs
Any forest dynamics model needs information on climate, vegetation
and soils of the forest stand to be simulated. Moreover, since models in
medfate differentiate between species, information on
species-specific model parameters is also needed. In this subsection we
explain the different steps to prepare the data needed to run function
fordyn().
Model inputs are explained in greater detail in vignettes Understanding
model inputs and Preparing
model inputs. Here we only review the different steps required
to run function fordyn().
Soil, vegetation, meteorology and species data
Soil information needs to be entered as a data frame
with soil layers in rows and physical attributes in columns. Soil
physical attributes can be initialized to default values, for a given
number of layers, using function defaultSoilParams():
examplesoil <- defaultSoilParams(4)
examplesoil## widths clay sand om nitrogen ph bd rfc
## 1 300 25 25 NA NA NA 1.5 25
## 2 700 25 25 NA NA NA 1.5 45
## 3 1000 25 25 NA NA NA 1.5 75
## 4 2000 25 25 NA NA NA 1.5 95As explained in the package overview, models included in
medfate were primarily designed to be ran on forest
inventory plots. Here we use the example object provided with
the package:
data(exampleforest)
exampleforest## $treeData
## Species DBH Height N Z50 Z95
## 1 Pinus halepensis 37.55 800 168 100 300
## 2 Quercus ilex 14.60 660 384 300 1000
##
## $shrubData
## Species Height Cover Z50 Z95
## 1 Quercus coccifera 80 3.75 200 1000
##
## attr(,"class")
## [1] "forest" "list"We can keep track of cohort age if we define a column called
Age in tree or shrub data, for example let us assume we
know the age of the two tree cohorts:
exampleforest$treeData$Age <- c(40, 24)Importantly, a data frame with daily weather for the period to be simulated is required. Here we use the default data frame included with the package:
## dates MinTemperature MaxTemperature Precipitation MinRelativeHumidity
## 1 2001-01-01 -0.5934215 6.287950 4.869109 65.15411
## 2 2001-01-02 -2.3662458 4.569737 2.498292 57.43761
## 3 2001-01-03 -3.8541036 2.661951 0.000000 58.77432
## 4 2001-01-04 -1.8744860 3.097705 5.796973 66.84256
## 5 2001-01-05 0.3288287 7.551532 1.884401 62.97656
## 6 2001-01-06 0.5461322 7.186784 13.359801 74.25754
## MaxRelativeHumidity Radiation WindSpeed
## 1 100.00000 12.89251 2.000000
## 2 94.71780 13.03079 7.662544
## 3 94.66823 16.90722 2.000000
## 4 95.80950 11.07275 2.000000
## 5 100.00000 13.45205 7.581347
## 6 100.00000 12.84841 6.570501Finally, simulations in medfate require a data frame
with species parameter values, which we load using defaults for
Catalonia (NE Spain):
data("SpParamsMED")Simulation control
Apart from data inputs, the behaviour of simulation models can be
controlled using a set of global parameters. The default
parameterization is obtained using function
defaultControl():
control <- defaultControl("Granier")Here we will run simulations of forest dynamics using the basic water
balance model (i.e. transpirationMode = "Granier"). The
complexity of the soil water balance calculations can be changed by
using "Sperry" as input to defaultControl().
However, when running fordyn() sub-daily output will never
be stored (i.e. setting subdailyResults = TRUE is
useless).
Executing the forest dynamics model
In this vignette we will fake a ten-year weather input by repeating the example weather data frame ten times.
meteo <- rbind(examplemeteo, examplemeteo, examplemeteo, examplemeteo,
examplemeteo, examplemeteo, examplemeteo, examplemeteo,
examplemeteo, examplemeteo)
meteo$dates <- as.character(seq(as.Date("2001-01-01"),
as.Date("2010-12-29"), by="day"))Now we run the forest dynamics model using all inputs (note that no
intermediate input object is needed, as in spwb() or
growth()):
fd<-fordyn(exampleforest, examplesoil, SpParamsMED, meteo, control,
latitude = 41.82592, elevation = 100)## Simulating year 2001 (1/10): (a) Growth/mortality, (b) Regeneration nT = 2 nS = 1
## Simulating year 2002 (2/10): (a) Growth/mortality, (b) Regeneration nT = 2 nS = 1
## Simulating year 2003 (3/10): (a) Growth/mortality, (b) Regeneration nT = 2 nS = 1
## Simulating year 2004 (4/10): (a) Growth/mortality, (b) Regeneration nT = 2 nS = 1
## Simulating year 2005 (5/10): (a) Growth/mortality, (b) Regeneration nT = 2 nS = 1
## Simulating year 2006 (6/10): (a) Growth/mortality, (b) Regeneration nT = 2 nS = 1
## Simulating year 2007 (7/10): (a) Growth/mortality, (b) Regeneration nT = 2 nS = 1
## Simulating year 2008 (8/10): (a) Growth/mortality, (b) Regeneration nT = 2 nS = 1
## Simulating year 2009 (9/10): (a) Growth/mortality, (b) Regeneration nT = 2 nS = 1
## Simulating year 2010 (10/10): (a) Growth/mortality, (b) Regeneration nT = 2 nS = 1It is worth noting that, while fordyn() calls function
growth() internally for each simulated year, the
verbose option of the control parameters only affects
function fordyn() (i.e. all console output from
growth() is hidden). Recruitment and summaries are done
only once a year at the level of function fordyn().
Inspecting model outputs
Stand, species and cohort summaries and plots
Among other outputs, function fordyn() calculates
standard summary statistics that describe the structural and
compositional state of the forest at each time step. For example, we can
access stand-level statistics using:
fd$StandSummary## Step NumTreeSpecies NumTreeCohorts NumShrubSpecies NumShrubCohorts
## 1 0 2 2 1 1
## 2 1 2 2 1 1
## 3 2 2 2 1 1
## 4 3 2 2 1 1
## 5 4 2 2 1 1
## 6 5 2 2 1 1
## 7 6 2 2 1 1
## 8 7 2 2 1 1
## 9 8 2 2 1 1
## 10 9 2 2 1 1
## 11 10 2 2 1 1
## TreeDensityLive TreeBasalAreaLive DominantTreeHeight DominantTreeDiameter
## 1 552.0000 25.03330 800.0000 37.55000
## 2 551.3665 25.19807 806.0378 37.66250
## 3 550.7277 25.36580 812.1627 37.77721
## 4 550.0833 25.53485 818.3165 37.89306
## 5 549.4316 25.70403 824.4565 38.00925
## 6 548.7762 25.87325 830.5686 38.12553
## 7 548.1152 26.04223 836.6478 38.24179
## 8 547.4486 26.21075 842.6854 38.35785
## 9 546.7746 26.37854 848.6801 38.47370
## 10 546.0968 26.54569 854.6307 38.58930
## 11 545.4173 26.71219 860.5372 38.70465
## QuadraticMeanTreeDiameter HartBeckingIndex ShrubCoverLive BasalAreaDead
## 1 24.02949 53.20353 3.750000 0.00000000
## 2 24.12229 52.83532 3.109453 0.03914817
## 3 24.21647 52.46727 3.202285 0.03978266
## 4 24.31126 52.10321 3.277538 0.04043173
## 5 24.40613 51.74584 3.374235 0.04120362
## 6 24.50095 51.39571 3.454956 0.04175781
## 7 24.59565 51.05302 3.552767 0.04243106
## 8 24.69012 50.71809 3.636510 0.04311020
## 9 24.78428 50.39086 3.739650 0.04391520
## 10 24.87810 50.07105 3.826331 0.04448469
## 11 24.97154 49.75834 3.932564 0.04492902
## ShrubCoverDead BasalAreaCut ShrubCoverCut
## 1 0.000000000 0 0
## 2 0.005314992 0 0
## 3 0.004835955 0 0
## 4 0.004968608 0 0
## 5 0.005110521 0 0
## 6 0.005235978 0 0
## 7 0.005369828 0 0
## 8 0.005512109 0 0
## 9 0.005667688 0 0
## 10 0.005801076 0 0
## 11 0.005912711 0 0Species-level analogous statistics are shown using:
fd$SpeciesSummary## Step Species NumCohorts TreeDensityLive TreeBasalAreaLive
## 1 0 Pinus halepensis 1 168.0000 18.604547
## 2 0 Quercus coccifera 1 NA NA
## 3 0 Quercus ilex 1 384.0000 6.428755
## 4 1 Pinus halepensis 1 167.6993 18.682696
## 5 1 Quercus coccifera 1 NA NA
## 6 1 Quercus ilex 1 383.6672 6.515376
## 7 2 Pinus halepensis 1 167.3960 18.762670
## 8 2 Quercus coccifera 1 NA NA
## 9 2 Quercus ilex 1 383.3317 6.603133
## 10 3 Pinus halepensis 1 167.0898 18.843401
## 11 3 Quercus coccifera 1 NA NA
## 12 3 Quercus ilex 1 382.9935 6.691452
## 13 4 Pinus halepensis 1 166.7801 18.923996
## 14 4 Quercus coccifera 1 NA NA
## 15 4 Quercus ilex 1 382.6515 6.780035
## 16 5 Pinus halepensis 1 166.4684 19.004374
## 17 5 Quercus coccifera 1 NA NA
## 18 5 Quercus ilex 1 382.3078 6.868877
## 19 6 Pinus halepensis 1 166.1540 19.084334
## 20 6 Quercus coccifera 1 NA NA
## 21 6 Quercus ilex 1 381.9612 6.957896
## 22 7 Pinus halepensis 1 165.8367 19.163697
## 23 7 Quercus coccifera 1 NA NA
## 24 7 Quercus ilex 1 381.6118 7.047055
## 25 8 Pinus halepensis 1 165.5159 19.242324
## 26 8 Quercus coccifera 1 NA NA
## 27 8 Quercus ilex 1 381.2587 7.136213
## 28 9 Pinus halepensis 1 165.1931 19.320377
## 29 9 Quercus coccifera 1 NA NA
## 30 9 Quercus ilex 1 380.9038 7.225314
## 31 10 Pinus halepensis 1 164.8693 19.397956
## 32 10 Quercus coccifera 1 NA NA
## 33 10 Quercus ilex 1 380.5480 7.314234
## ShrubCoverLive BasalAreaDead ShrubCoverDead BasalAreaCut ShrubCoverCut
## 1 NA 0.000000000 NA 0 NA
## 2 3.750000 NA 0.000000000 NA 0
## 3 NA 0.000000000 NA 0 NA
## 4 NA 0.033496809 NA 0 NA
## 5 3.109453 NA 0.005314992 NA 0
## 6 NA 0.005651361 NA 0 NA
## 7 NA 0.034003668 NA 0 NA
## 8 3.202285 NA 0.004835955 NA 0
## 9 NA 0.005778989 NA 0 NA
## 10 NA 0.034522299 NA 0 NA
## 11 3.277538 NA 0.004968608 NA 0
## 12 NA 0.005909431 NA 0 NA
## 13 NA 0.035144929 NA 0 NA
## 14 3.374235 NA 0.005110521 NA 0
## 15 NA 0.006058690 NA 0 NA
## 16 NA 0.035581012 NA 0 NA
## 17 3.454956 NA 0.005235978 NA 0
## 18 NA 0.006176799 NA 0 NA
## 19 NA 0.036117782 NA 0 NA
## 20 3.552767 NA 0.005369828 NA 0
## 21 NA 0.006313276 NA 0 NA
## 22 NA 0.036658673 NA 0 NA
## 23 3.636510 NA 0.005512109 NA 0
## 24 NA 0.006451523 NA 0 NA
## 25 NA 0.037305675 NA 0 NA
## 26 3.739650 NA 0.005667688 NA 0
## 27 NA 0.006609523 NA 0 NA
## 28 NA 0.037751831 NA 0 NA
## 29 3.826331 NA 0.005801076 NA 0
## 30 NA 0.006732854 NA 0 NA
## 31 NA 0.038091392 NA 0 NA
## 32 3.932564 NA 0.005912711 NA 0
## 33 NA 0.006837627 NA 0 NAPackage medfate provides a simple plot
function for objects of class fordyn. For example, we can
show the interannual variation in stand-level basal area using:
plot(fd, type = "StandBasalArea")
Tree/shrub tables
Another useful output of fordyn() are tables in long
format with cohort structural information (i.e. DBH, height, density,
etc) for each time step:
fd$TreeTable## Step Year Cohort Species DBH Height N Z50 Z95 Z100
## 1 0 NA T1_148 Pinus halepensis 37.55000 800.0000 168.0000 100 300 NA
## 2 0 NA T2_168 Quercus ilex 14.60000 660.0000 384.0000 300 1000 NA
## 3 1 2001 T1_148 Pinus halepensis 37.66250 806.0378 167.6993 100 300 NA
## 4 1 2001 T2_168 Quercus ilex 14.70440 663.2431 383.6672 300 1000 NA
## 5 2 2002 T1_148 Pinus halepensis 37.77721 812.1627 167.3960 100 300 NA
## 6 2 2002 T2_168 Quercus ilex 14.80958 666.5015 383.3317 300 1000 NA
## 7 3 2003 T1_148 Pinus halepensis 37.89306 818.3165 167.0898 100 300 NA
## 8 3 2003 T2_168 Quercus ilex 14.91487 669.7555 382.9935 300 1000 NA
## 9 4 2004 T1_148 Pinus halepensis 38.00925 824.4565 166.7801 100 300 NA
## 10 4 2004 T2_168 Quercus ilex 15.01998 672.9955 382.6515 300 1000 NA
## 11 5 2005 T1_148 Pinus halepensis 38.12553 830.5686 166.4684 100 300 NA
## 12 5 2005 T2_168 Quercus ilex 15.12486 676.2206 382.3078 300 1000 NA
## 13 6 2006 T1_148 Pinus halepensis 38.24179 836.6478 166.1540 100 300 NA
## 14 6 2006 T2_168 Quercus ilex 15.22946 679.4285 381.9612 300 1000 NA
## 15 7 2007 T1_148 Pinus halepensis 38.35785 842.6854 165.8367 100 300 NA
## 16 7 2007 T2_168 Quercus ilex 15.33373 682.6182 381.6118 300 1000 NA
## 17 8 2008 T1_148 Pinus halepensis 38.47370 848.6801 165.5159 100 300 NA
## 18 8 2008 T2_168 Quercus ilex 15.43757 685.7861 381.2587 300 1000 NA
## 19 9 2009 T1_148 Pinus halepensis 38.58930 854.6307 165.1931 100 300 NA
## 20 9 2009 T2_168 Quercus ilex 15.54089 688.9294 380.9038 300 1000 NA
## 21 10 2010 T1_148 Pinus halepensis 38.70465 860.5372 164.8693 100 300 NA
## 22 10 2010 T2_168 Quercus ilex 15.64353 692.0438 380.5480 300 1000 NA
## Age ObsID
## 1 40 <NA>
## 2 24 <NA>
## 3 40 NA
## 4 24 NA
## 5 41 NA
## 6 25 NA
## 7 42 NA
## 8 26 NA
## 9 43 NA
## 10 27 NA
## 11 44 NA
## 12 28 NA
## 13 45 NA
## 14 29 NA
## 15 46 NA
## 16 30 NA
## 17 47 NA
## 18 31 NA
## 19 48 NA
## 20 32 NA
## 21 49 NA
## 22 33 NAThe same can be shown for dead trees:
fd$DeadTreeTable## Step Year Cohort Species DBH Height N N_starvation
## 1 1 2001 T1_148 Pinus halepensis 37.66250 806.0378 0.3006735 0
## 2 1 2001 T2_168 Quercus ilex 14.70440 663.2431 0.3327885 0
## 3 2 2002 T1_148 Pinus halepensis 37.77721 812.1627 0.3033724 0
## 4 2 2002 T2_168 Quercus ilex 14.80958 666.5015 0.3354877 0
## 5 3 2003 T1_148 Pinus halepensis 37.89306 818.3165 0.3061191 0
## 6 3 2003 T2_168 Quercus ilex 14.91487 669.7555 0.3382336 0
## 7 4 2004 T1_148 Pinus halepensis 38.00925 824.4565 0.3097377 0
## 8 4 2004 T2_168 Quercus ilex 15.01998 672.9955 0.3419403 0
## 9 5 2005 T1_148 Pinus halepensis 38.12553 830.5686 0.3116712 0
## 10 5 2005 T2_168 Quercus ilex 15.12486 676.2206 0.3437881 0
## 11 6 2006 T1_148 Pinus halepensis 38.24179 836.6478 0.3144523 0
## 12 6 2006 T2_168 Quercus ilex 15.22946 679.4285 0.3465741 0
## 13 7 2007 T1_148 Pinus halepensis 38.35785 842.6854 0.3172329 0
## 14 7 2007 T2_168 Quercus ilex 15.33373 682.6182 0.3493626 0
## 15 8 2008 T1_148 Pinus halepensis 38.47370 848.6801 0.3208906 0
## 16 8 2008 T2_168 Quercus ilex 15.43757 685.7861 0.3531198 0
## 17 9 2009 T1_148 Pinus halepensis 38.58930 854.6307 0.3227857 0
## 18 9 2009 T2_168 Quercus ilex 15.54089 688.9294 0.3549423 0
## 19 10 2010 T1_148 Pinus halepensis 38.70465 860.5372 0.3237507 0
## 20 10 2010 T2_168 Quercus ilex 15.64353 692.0438 0.3557509 0
## N_dessication N_burnt N_resprouting_stumps Z50 Z95 Z100 Age ObsID
## 1 0 0 0 100 300 NA 40 NA
## 2 0 0 0 300 1000 NA 24 NA
## 3 0 0 0 100 300 NA 40 NA
## 4 0 0 0 300 1000 NA 24 NA
## 5 0 0 0 100 300 NA 41 NA
## 6 0 0 0 300 1000 NA 25 NA
## 7 0 0 0 100 300 NA 42 NA
## 8 0 0 0 300 1000 NA 26 NA
## 9 0 0 0 100 300 NA 43 NA
## 10 0 0 0 300 1000 NA 27 NA
## 11 0 0 0 100 300 NA 44 NA
## 12 0 0 0 300 1000 NA 28 NA
## 13 0 0 0 100 300 NA 45 NA
## 14 0 0 0 300 1000 NA 29 NA
## 15 0 0 0 100 300 NA 46 NA
## 16 0 0 0 300 1000 NA 30 NA
## 17 0 0 0 100 300 NA 47 NA
## 18 0 0 0 300 1000 NA 31 NA
## 19 0 0 0 100 300 NA 48 NA
## 20 0 0 0 300 1000 NA 32 NAAccessing the output from function growth()
Since function fordyn() makes internal calls to function
growth(), it stores the result in a vector called
GrowthResults, which we can use to inspect intra-annual
patterns of desired variables. For example, the following shows the leaf
area for individuals of the three cohorts during the second year:
plot(fd$GrowthResults[[2]], "LeafArea", bySpecies = T)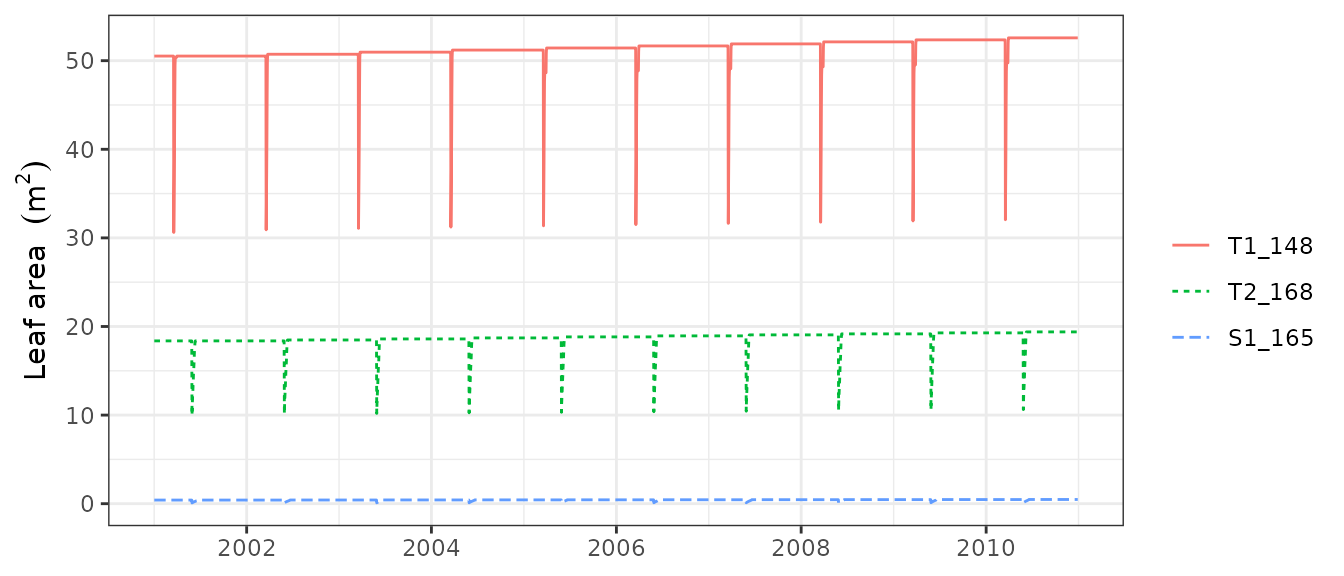 Instead of examining year by year, it is possible to plot the whole
series of results by passing a
Instead of examining year by year, it is possible to plot the whole
series of results by passing a fordyn object to the
plot() function:
plot(fd, "LeafArea")
We can also create interactive plots for particular steps using
function shinyplot(), e.g.:
shinyplot(fd$GrowthResults[[1]])Finally, calling function extract() will extract and
bind outputs for all the internal calls to function
growth():
## # A tibble: 3,650 × 51
## date PET Precipitation Rain Snow NetRain Snowmelt Infiltration
## <date> [L/m^2] [L/m^2] [L/m^2] [L/m^… [L/m^2] [L/m^2] [L/m^2]
## 1 2001-01-01 0.883 4.87 4.87 0 3.60 0 3.60
## 2 2001-01-02 1.64 2.50 2.50 0 1.25 0 1.25
## 3 2001-01-03 1.30 0 0 0 0 0 0
## 4 2001-01-04 0.569 5.80 5.80 0 4.54 0 4.54
## 5 2001-01-05 1.68 1.88 1.88 0 0.822 0 0.822
## 6 2001-01-06 1.21 13.4 13.4 0 11.9 0 11.9
## 7 2001-01-07 0.637 5.38 0 5.38 0 0 0
## 8 2001-01-08 0.832 0 0 0 0 0 0
## 9 2001-01-09 1.98 0 0 0 0 0 0
## 10 2001-01-10 0.829 5.12 5.12 0 3.85 5.38 9.23
## # ℹ 3,640 more rows
## # ℹ 43 more variables: InfiltrationExcess [L/m^2], SaturationExcess [L/m^2],
## # Runoff [L/m^2], DeepDrainage [L/m^2], CapillarityRise [L/m^2],
## # Evapotranspiration [L/m^2], Interception [L/m^2], SoilEvaporation [L/m^2],
## # HerbTranspiration [L/m^2], PlantExtraction [L/m^2], Transpiration [L/m^2],
## # HydraulicRedistribution [L/m^2], LAI [m^2/m^2], LAIherb [m^2/m^2],
## # LAIlive [m^2/m^2], LAIexpanded [m^2/m^2], LAIdead [m^2/m^2], Cm [L/m^2], …Forest dynamics including management
The package allows including forest management in simulations of
forest dynamics. This is done in a very flexible manner, in the sense
that fordyn() allows the user to supply an arbitrary
function implementing a desired management strategy for the stand whose
dynamics are to be simulated. The package includes, however, an in-built
default function called defaultManagementFunction() along
with a flexible parameterization, a list with defaults provided by
function defaultManagementArguments().
Here we provide an example of simulations including forest management:
# Default arguments
args <- defaultManagementArguments()
# Here one can modify defaults before calling fordyn()
#
# Simulation
fd<-fordyn(exampleforest, examplesoil, SpParamsMED, meteo, control,
latitude = 41.82592, elevation = 100,
management_function = defaultManagementFunction,
management_args = args)## Simulating year 2001 (1/10): (a) Growth/mortality & management [thinning], (b) Regeneration nT = 2 nS = 2
## Simulating year 2002 (2/10): (a) Growth/mortality & management [none], (b) Regeneration nT = 2 nS = 2
## Simulating year 2003 (3/10): (a) Growth/mortality & management [none], (b) Regeneration nT = 2 nS = 2
## Simulating year 2004 (4/10): (a) Growth/mortality & management [none], (b) Regeneration nT = 2 nS = 2
## Simulating year 2005 (5/10): (a) Growth/mortality & management [none], (b) Regeneration nT = 2 nS = 2
## Simulating year 2006 (6/10): (a) Growth/mortality & management [none], (b) Regeneration nT = 2 nS = 2
## Simulating year 2007 (7/10): (a) Growth/mortality & management [none], (b) Regeneration nT = 2 nS = 2
## Simulating year 2008 (8/10): (a) Growth/mortality & management [none], (b) Regeneration nT = 2 nS = 2
## Simulating year 2009 (9/10): (a) Growth/mortality & management [none], (b) Regeneration nT = 2 nS = 2
## Simulating year 2010 (10/10): (a) Growth/mortality & management [none], (b) Regeneration nT = 2 nS = 2When management is included in simulations, two additional tables are produced, corresponding to the trees and shrubs that were cut, e.g.:
fd$CutTreeTable## Step Year Cohort Species DBH Height N Z50 Z95 Z100
## 1 1 2001 T1_148 Pinus halepensis 37.6625 806.0378 9.371556 100 300 NA
## 2 1 2001 T2_168 Quercus ilex 14.7044 663.2431 383.667212 300 1000 NA
## Age ObsID
## 1 40 NA
## 2 24 NAManagement parameters were those of an irregular model with thinning interventions from ‘below’, indicating that smaller trees were to be cut earlier:
args$type## [1] "irregular"
args$thinning## [1] "below"Note that in this example, there is resprouting of Quercus ilex after the thinning intervention, evidenced by the new cohort (T3_168) appearing in year 2001:
fd$TreeTable## Step Year Cohort Species DBH Height N Z50
## 1 0 NA T1_148 Pinus halepensis 37.550000 800.00000 168.0000 100.0000
## 2 0 NA T2_168 Quercus ilex 14.600000 660.00000 384.0000 300.0000
## 3 1 2001 T1_148 Pinus halepensis 37.662500 806.03781 158.3278 100.0000
## 4 1 2001 T3_168 Quercus ilex 1.000000 47.23629 3000.0000 300.0000
## 5 2 2002 T1_148 Pinus halepensis 37.778853 812.18467 158.1566 100.0000
## 6 2 2002 T3_168 Quercus ilex 1.018293 48.34502 2942.7311 300.0000
## 7 3 2003 T1_148 Pinus halepensis 37.898467 818.47725 157.9844 100.0000
## 8 3 2003 T3_168 Quercus ilex 1.034482 49.33046 2893.8003 300.0000
## 9 4 2004 T1_148 Pinus halepensis 38.018498 824.75803 157.8106 100.0000
## 10 4 2004 T3_168 Quercus ilex 1.050840 50.32717 2845.9321 300.0000
## 11 5 2005 T1_148 Pinus halepensis 38.138587 831.00805 157.6362 100.0000
## 12 5 2005 T3_168 Quercus ilex 1.067521 51.34412 2798.6803 300.0000
## 13 6 2006 T1_148 Pinus halepensis 38.258610 837.22082 157.4607 100.0000
## 14 6 2006 T3_168 Quercus ilex 1.084579 52.38464 2751.9084 300.0000
## 15 7 2007 T1_148 Pinus halepensis 38.378576 843.39721 157.2841 100.0000
## 16 7 2007 T3_168 Quercus ilex 1.102018 53.44895 2705.6376 300.0000
## 17 8 2008 T1_148 Pinus halepensis 38.498366 849.53123 157.1059 100.0000
## 18 8 2008 T3_168 Quercus ilex 1.119841 54.53728 2659.8788 300.0000
## 19 9 2009 T1_148 Pinus halepensis 38.617954 855.62195 156.9271 100.0000
## 20 9 2009 T2_168 Quercus ilex 1.120493 54.74958 3020.6418 314.9779
## 21 10 2010 T1_148 Pinus halepensis 38.733859 861.49404 156.7483 100.0000
## 22 10 2010 T2_168 Quercus ilex 1.138176 55.82883 2614.3610 314.9779
## Z95 Z100 Age ObsID
## 1 300.000 NA 40.00000 <NA>
## 2 1000.000 NA 24.00000 <NA>
## 3 300.000 NA 40.00000 NA
## 4 1000.000 NA 24.00000 <NA>
## 5 300.000 NA 41.00000 NA
## 6 1000.000 NA 24.00000 NA
## 7 300.000 NA 42.00000 NA
## 8 1000.000 NA 25.00000 NA
## 9 300.000 NA 43.00000 NA
## 10 1000.000 NA 26.00000 NA
## 11 300.000 NA 44.00000 NA
## 12 1000.000 NA 27.00000 NA
## 13 300.000 NA 45.00000 NA
## 14 1000.000 NA 28.00000 NA
## 15 300.000 NA 46.00000 NA
## 16 1000.000 NA 29.00000 NA
## 17 300.000 NA 47.00000 NA
## 18 1000.000 NA 30.00000 NA
## 19 300.000 NA 49.00000 <NA>
## 20 1430.369 NA 29.10946 <NA>
## 21 300.000 NA 49.00000 NA
## 22 1430.369 NA 29.10946 NAReferences
- De Cáceres M, Molowny-Horas R, Cabon A, Martínez-Vilalta J, Mencuccini M, García-Valdés R, Nadal-Sala D, Sabaté S, Martin-StPaul N, Morin X, D’Adamo F, Batllori E, Améztegui A (2023) MEDFATE 2.9.3: A trait-enabled model to simulate Mediterranean forest function and dynamics at regional scales. Geoscientific Model Development 16: 3165-3201 (https://doi.org/10.5194/gmd-16-3165-2023).