Chapter 1 Introduction
This chapter provides an overview of the purpose of developing the medfate an medfateland R packages, their main simulation functions and expected applications.
1.1 Purpose
Being able to anticipate the impact of global change on forest function and dynamics is one of the major environmental challenges in contemporary societies. However, uncertainties in the mechanisms underpinning forest function and practical constraints in how to integrate available information still hinder the availability of robust and reliable predictive models. Despite the amount of knowledge accumulated about the function and dynamics of forest ecosystems and the plethora of observations and modelling tools already available, further efforts are necessary to achieve the challenge of integrating different global change drivers into simulation frameworks useful for research and applications.
1.1.1 Package medfate
The R package medfate has been designed as a platform to simulate the function and dynamics of individual forest stands at temporal scales from days to years. Climate is the main environmental driver forcing model predictions, and the ecological, biophysical, hydrological and physiological processes surrounding these are fundamental for the simulation models included in the package. In particular, the package allows the simulation of daily energy, water and carbon balances within forest stands. It also allows simulating the growth, mortality and regeneration processes of a set of plant cohorts competing for light and water within a forest stand. Finally, additional package functions allow relating the amount of plant biomass and the water status of plant live and dead tissues to fuel characteristics and, hence, fire hazard.
The version of the reference manual that you are reading is intended to reflect version 5.0.0 of the package.
1.1.2 Package medfateland
The R package medfateland has been designed to extend the capabilities of medfate to a spatial context, most usually at the landscape or regional level. It allows running the stand-level models available in medfate on a set of spatial units (normally points or cells) within a target area. The package allows coordinating the dynamics of multiple stands via the evaluation of demand-based management scenarios. Additionally, the package allows considering lateral water transfer processes of forested watersheds. Hence, medfateland can also be used as a tool for eco-hydrological applications.
The version of the reference manual that you are reading is intended to reflect version 3.0.0 of the package.
1.1.3 Companion packages
Packages medfate and medfateland are more easily used in conjunction with three other packages, called meteoland, traits4models and forestables. Package meteoland assists the estimation of weather variables over landscapes, whereas traits4models provides functions to assist the harmonization of plant trait data bases with the aim to create species parameter tables for simulation models, and forestables allows reading and harmonizing forest inventory data. Together the five R packages conform a modelling framework designed to help simulating the function and dynamics of forest ecosystems, with a particular focus on drought impacts under Mediterranean conditions (Fig. 1.1).
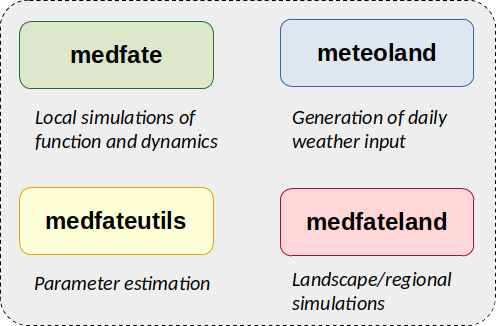
Figure 1.1: Set of packages conforming the medfate modelling framework. Black arrows indicate library requirements, whereas gray arrows indicate model parameter or data provision.
Although meteoland and forestables are part of the forest modelling framework, they can be used independently for many other purposes. Similarly, package traits4models can be used to generate parameter inputs for other models or frameworks.
1.2 Installation
1.2.1 Requirements
Both medfate and medfateland have a user interface in R, but models have been coded in C++ language, and the resulting simulation functions are linked to R via the Rcpp package. This means that whenever installation is done from sources you will need a C++ compiler (i.e. Rtools in Windows). Note that compilers will always be a requirement when installing packages from GitHub.
1.2.2 Installing medfate
Package medfate can be found at CRAN, where it is updated every few months. Hence, it can be installed as any other R package using:
Users can also download and install the latest stable versions GitHub as follows (required package remotes should be installed/updated first):
Among other dependencies, installing medfate requires package meteoland, as it links to some of its C++ functions. Note that some package dependencies are only suggested, so that they are not automatically installed when installing medfate.
1.2.3 Installing medfateland
Since both packages evolve together, installing medfateland normally requires an up-to-date version of medfate. Package medfateland can be found at CRAN, where it is updated every few months. Hence, it can be installed as any other R package using:
Users can also download and install the latest stable versions GitHub as follows:
Among other dependencies, installing medfateland requires packages medfate and meteoland, as it links to some of their C++ functions.
1.3 Main functions in medfate
1.3.1 Dynamic simulation functions
Dynamic simulation functions in medfate include three nested simulation levels (Fig. 1.2):
- Forest water and energy balance can be studied for a given forest stand using function
spwb()(soil-plant-water-balance). The same function is used to run three different models (Granier,SperryorSureau), that correspond to two levels of complexity (basic or advanced; see Fig. 1.2). The user should choose one model or another depending on the intended application and data availability. Water balance simulations include hydrological processes (rainfall interception, soil infiltration, percolation and evapotranspiration) and plant physiological processes related to transpiration (hydraulics, photosynthesis and stomatal regulation). Most processes are implemented at the daily scale, although some operate at subdaily time steps for advanced models. Functionspwb()has a closely related functionspwb_day()that allows single-day simulation. - Changes in primary (leaf area) and secondary (sapwood) growth are key to evaluate the influence of climatic conditions on plant and forest structure and function. Function
growth()extends the previous models because it allows simulating carbon balance, tissue growth and mortality of a set of plant cohorts competing for light and water in a given forest stand. Carbon balance is performed at the plant level, at the cohort level (once mortality is included), and at the stand level, after accounting for heterotrophic respiration due to decomposition processes. Analogously to the simulation of water balance,growth()has a closely related functiongrowth_day()that allows focusing on sub-daily processes (i.e. carbon balance). - Finally, function
fordyn()complements growth and mortality processes with recruitment (from seeds) and resprouting, thus completing the minimum set of demographic processes needed to simulate Mediterranean forest dynamics. The function splits the period to be simulated by years and makes internal calls togrowth()for the simulation of growth and mortality.fordyn()is suited to simulate the inter-annual variation in forest structure and composition, while accounting for the biophysical and physiological processes provided by the former models. In addition, in allows simulating human interventions (i.e. tree or shrub cutting or planting) on forest stands.
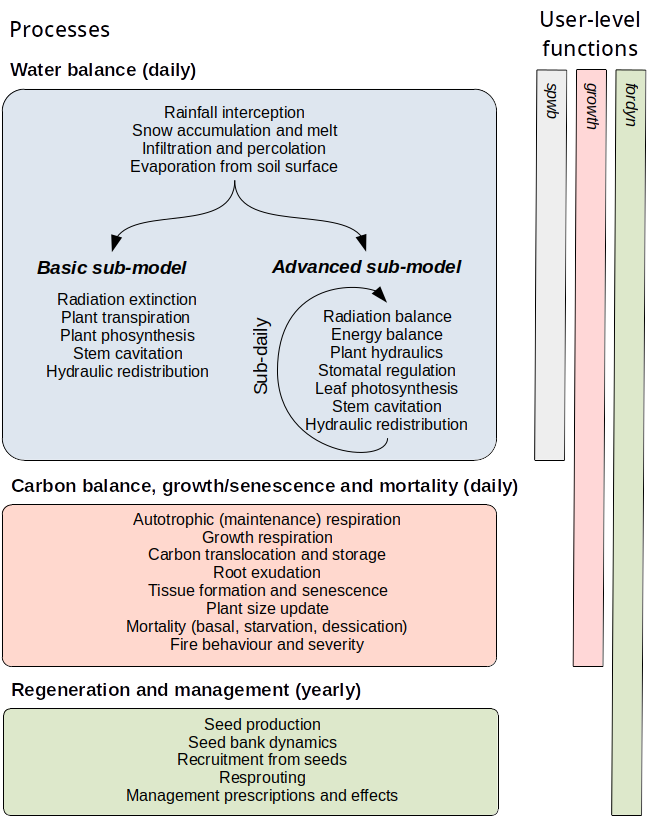
Figure 1.2: Nested relationships between medfate simulation functions and processes included in each model
1.3.2 Sub-model functions
Many of the sub-models included in medfate are implemented as C++ functions and internally called by the simulation functions implementing top-level models. Most sub-models have, however, their corresponding R function to made them directly available to the user. This facilitates understanding the different sub-models and a more creative use of the package. However, most users will not normally use them in their simulation workflows. Sub-model functions are grouped by subject, which is included in the name of the function. The different sub-model functions are (by subject):
| Group | Description |
|---|---|
biophysics_* |
Physics and biophysics |
carbon_* |
Biomass estimation and carbon balance |
decomposition_* |
Dynamics of litter and soil organic carbon pools |
fuel_* |
Fuel properties |
fire_* |
Fire behaviour and severity |
hydraulics_* |
Plant hydraulics |
hydrology_* |
Canopy and soil hydrology |
light_* |
Light extinction and absortion |
moisture_* |
Live tissue moisture |
mortality_* |
Plant mortality |
pheno_* |
Leaf phenology |
photo_* |
Leaf photosynthesis |
regeneration_* |
Regeneration from seed germination or resprouting |
root_* |
Root distribution and conductance calculations |
soil_* |
Soil hydraulics and thermodynamics |
transp_* |
Stomatal regulation, transpiration and photosynthesis |
wind_* |
Canopy turbulence |
1.3.3 Static functions
Package medfate includes a number of functions to examine static properties of plants conforming forest stands, summary functions at the species- and stand- levels:
plant_*: Cohort-level information (species name, id, leaf area, height, etc.).species_*: Cohort-level attributes aggregated by species (e.g. basal area, cover, density, etc.).herb_*: Attributes of the (aggregated) herbaceous layer (e.g. fuel density or leaf area index).stand_*: Stand-level attributes (e.g. stand basal area, fuel loading, etc.).
Vegetation functioning and dynamics have strong, but complex, effects on fire hazard. On one hand, growth and death of organs and individuals changes the amount of standing live and dead fuels, as well as downed dead fuels. On the other, day-to-day changes in soil and plant water content changes the physical properties of fuel, notably moisture content of live and dead fuels. Package medfate provides functions to estimate fuel properties and potential fire behaviour in forest stands. Specifically, fuel_FCCS() calculates fuel characteristics for a set of fuel strata; and a fire behaviour model is implemented in function fire_FCCS() to calculate the intensity of surface fire reaction and the rate of fire spread of surface fires assuming a steady-state fire. Fuel and fire behavior functions allow obtaining the following:
- Fuel characteristics by stratum.
- Surface fire behavior (i.e. reaction intensity, rate of spread, fireline intensity and flame length).
- Crown fire behavior.
- Fire potential ratings of surface fire behavior and crown fire behavior.
1.4 Main functions in medfateland
Package medfateland allows simulating forest functioning and dynamics on sets forests stands distributed across space, with or without spatial processes (Fig. 1.3).
When spatial processes are omitted, the package offers a set of simulation functions that are analogous to those of package medfate but allow processing multiple forest stands sequentially or in parallel:
- Forest water and energy balance can be studied for a set of forest stands using functions
spwb_spatial()orspwb_spatial_day(). - Growth and mortality of a set of plant cohorts competing for light and water can be simulated for a set of forest stands using functions
growth_spatial()orgrowth_spatial_day(). - Forest dynamics arising from competition for light and water can be simulated for a set of forest stands using function
fordyn_spatial().
The package offers a simulation function where the dynamics of forest stands are related by management decisions:
- Function
fordyn_scenario()allows simulatingfordyn()on forest stands, while applying a demand-based forest management scenario. Simulations with this function also include seed dispersal among forest stands.
Finally, there are three functions that are meant to simulate watersheds, including spatial processes:
- Functions
spwb_land()andgrowth_land()include distributed hydrological models that allow simulating daily water balance or growth processes on the cells of a watershed while accounting for overland runoff, subsurface flow and groundwater flow between cells. - Forest dynamics arising from competition for light and water can be simulated in a watershed using function
fordyn_land(). Simulations with this function also include seed dispersal among grid cells.
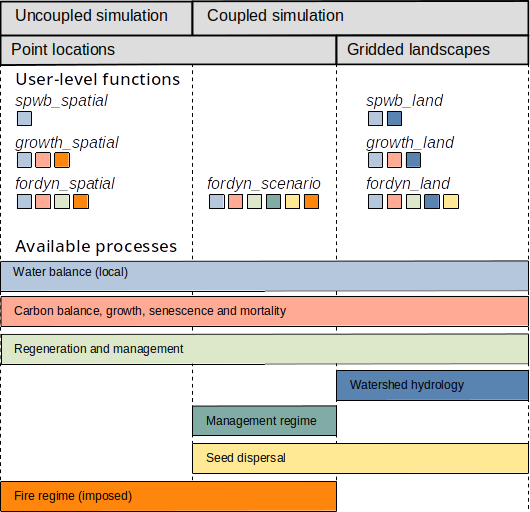
Figure 1.3: Overview of simulation functions in medfateland and the processes available for each of them. Colored squares below each simulation function indicate the processes included